RNA and why we love it
The cornerstone of our lab is RNA: we are fascinated by the functional diversity of this molecule and its importance for the cell today and during the evolution of life.
Currently, we assume that the earliest moment in molecular evolution that allowed for heredity is represented by an “RNA world”. This term describes an environment, where genetic information was passed on in the form of RNA, which also acted as the key catalyst, and highlights the unique role of RNA in the history of our existence. Nowadays, RNA has generally lost the function of information storage to DNA and the function of catalysis to proteins. Nevertheless, RNA is still critical for all cellular functions. It acts during multiple steps in the translation of genetic information into proteins. Furthermore, RNA can act as a ribozyme and through a plethora of small and long non-coding RNA (ncRNA) regulates gene expression and protein translation.
It is not surprising that the extremely diverse field of RNA biology has attracted increasing attention in recent years, leading to exciting new discoveries, which constantly continuously update our view of numerous cellular processes and their regulation. In the Leidel lab, we combine classical methods of molecular biology and biochemistry with modern sequencing technologies to study RNA and the role of various RNA species in translation – a crucial cellular process essential to building and maintaining life.
Our experimental models

Our methods
RNA modifications
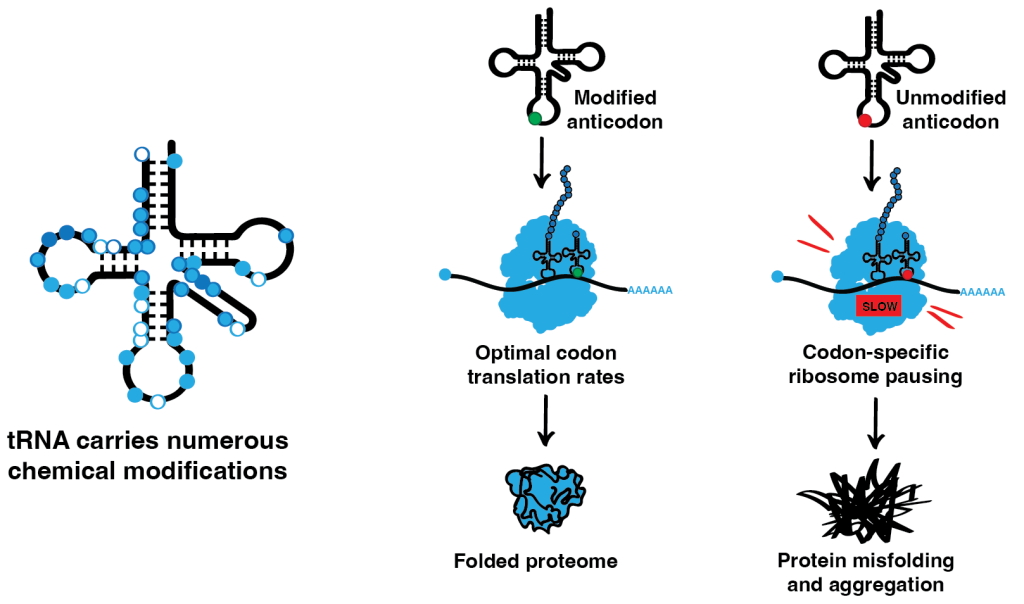
Most RNA species carry numerous posttranscriptional chemical modifications, which regulate their function and/or stability. One of our key research directions looks deeply into the role of posttranscriptional modifications of tRNA, the most heavily modified RNA species. tRNAs are major players in translation. They act as adaptor molecules, physically connecting the triplets of the genetic code to the corresponding amino acids. The modifications in the tRNA anticodon loop are thought to be critical for codon-anticodon interaction during translation, while the modifications in the tRNA body mainly affect tRNA folding and stability. However, the exact mechanisms and in vivo roles of numerous tRNA modifications are not fully understood. In our lab, we investigate the roles of tRNA modifications in different model organisms and use ribosome profiling in combination with other advanced sequencing techniques to analyze how the absence of tRNA modifications influences translation dynamics at the codon level. We complement our experiments with a comprehensive analysis of the cellular proteome to investigate the effects that these codon-specific changes in translation have on protein homeostasis.
In addition to tRNA, we focus on mRNA modifications since both types of modifications can affect codon-anticodon interactions. We are particularly interested in m6A, the most abundant internal mRNA modification. m6A modification affects various steps of mRNA metabolism, including splicing, export, translation, and mRNA stability, and as a result, is critical for multiple cellular processes. Finally, we investigate noncanonical mRNA capping to understand the cellular consequences of perturbation of noncanonical mRNA cap turnover.
Small peptides
Ribosome profiling is an excellent tool for gene discovery. The method allowed us to discover actively translated short open reading frames (ORFs) on transcripts, which were previously annotated as non-coding. After confirming the presence of these short peptides in vivo, we characterize their roles in cellular homeostasis and vertebrate development.
Translational quality control
The process of protein synthesis is incredibly intricate and complex and requires a plethora of players: mRNAs, ribosomes, tRNAs, initiation-, elongation-, and termination factors. Every member of this “translation team” can alter the process or even lead to its complete failure: e.g., mRNA may contain defective nucleotides or a premature stop codon, ribosomes may be defective and inefficient at decoding the mRNA sequence, tRNAs may lack a crucial chemical modification or their expression levels may be altered. Since protein synthesis is essential to life, cells have evolved multiple quality-control systems to either avoid or remedy such perturbations of mRNA translation. Importantly, mutations that affect translation dynamics or factors implicated in quality control are often linked to human degenerative diseases.
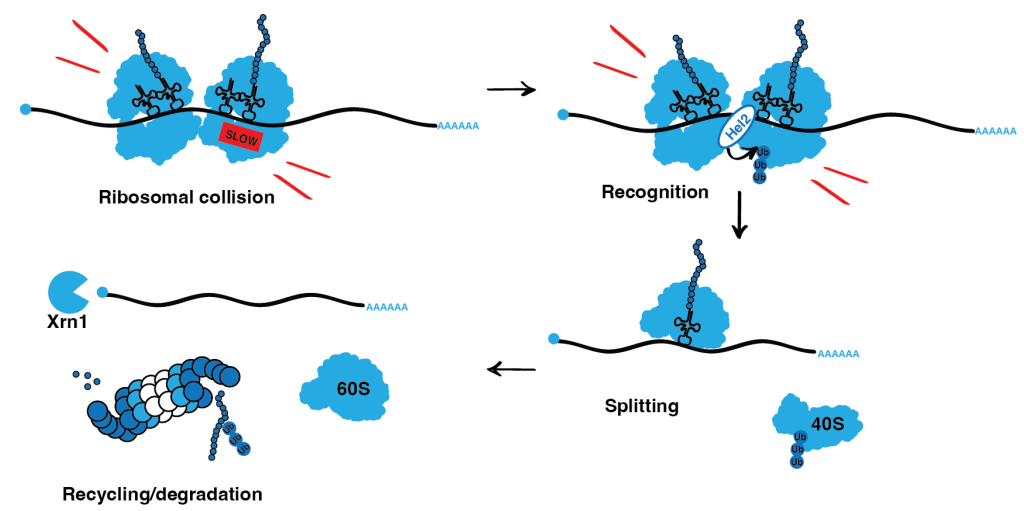
As ribosomes move along mRNA, the newly synthesized peptides need to be protected from aggregation to fold into a functional protein. We found that codon-specific translation defects can lead to the aggregation of cellular proteins, a hallmark of human degenerative diseases. But how are translation dynamics mechanistically linked to accurate cotranslational folding?
Ribosomes can slow down while decoding a particular codon on mRNA. If such a slowdown is not resolved in time and a ribosome does not continue its movement, the trailing ribosome can catch up and collide with the slow ribosome. Members of the Ribosome-associated Quality Control (RQC) pathway recognize the interface of collided ribosomes and resolve the situation by cleaving the mRNA, splitting and recycling the ribosomes while at the same time targeting the nascent peptide is degraded.
We are particularly interested in the consequences of defective RQC system and co-translational chaperones on the proteome of the cell and how these quality control mechanisms contribute to the way the cell deals with codon-specific translational stress.
Multiomics: integrating dynamic analyses of the transcriptome, translatome, and proteome
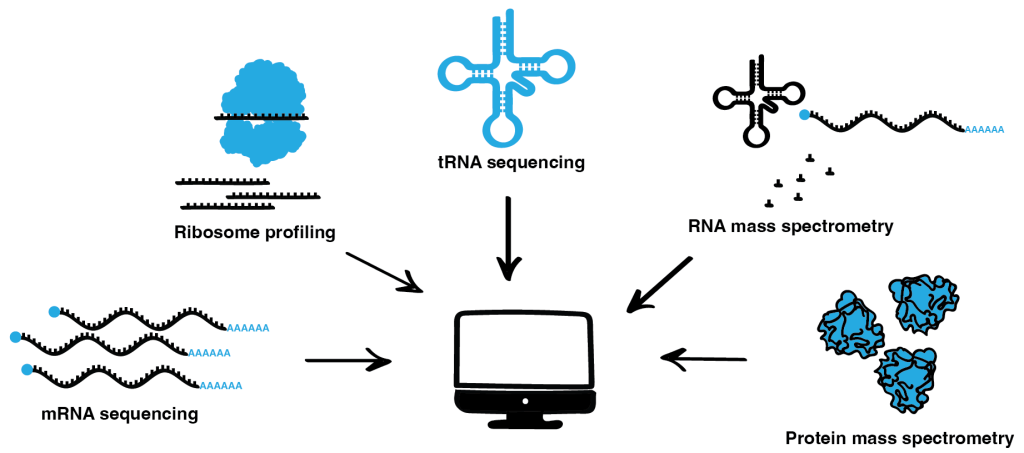
Over the years, we have diversified our portfolio of omics methods. We perform high-throughput and high-resolution ribosome profiling, RNAseq, tRNAseq, and protein and RNA mass spectrometry. Furthermore, we constantly continue to refine these main experimental techniques in our lab to add new state-of-the-art methods to our toolkit. Using these quality data allows us to assess changes at all steps of gene expression and protein synthesis: mRNA/tRNA transcription and modification, translation efficiency with codon resolution, and the quality and abundance of newly synthesized proteins. Here, the computational and experimental members of our group work hand-in-hand to integrate these multilevel data to unravel the mechanistic links between the various cellular processes to gain a comprehensive understanding of how translation dynamics influence phenotypes.